Synteny of MHC Paralogon
The conservation of gene synteny on different chromosomal regions entails the functional significance of these genes. For instance, the co-expression of neighboring genes is mediated by high order structural organization of chromosomes, which brings together the far off genomic regions in close proximity in order to be expressed together in a coordinated manner (Parveen et al. 2013). Similarly, the gene regulatory elements spread across long regions impose critical constraints on genomic architecture and are known to have maintained exceptionally long syntenic blocks both within and across species (Ali et al., 2016).
Gene families with threefold or fourfold representation in the proposed paralogy regions on human chromosomes 1, 6, 9 and 19 (MHC-bearing chromosomes) were identified by scanning the human genome sequence maps available at the Ensembl and UCSC genome browsers (Flicek et al., 2011; Fujita et al., 2011; Hubbard et al., 2002). A total of 40 gene families with at least three fold representations on these chromosomes were identified. Schematic synteny map of these 40 multigene families was manually sketched to depict their relative positioning on respective chromosomal locations. Members of these gene families also have representations on non-MHC bearing chromosomes. Such members are not drawn on synteny map (Figure 1).
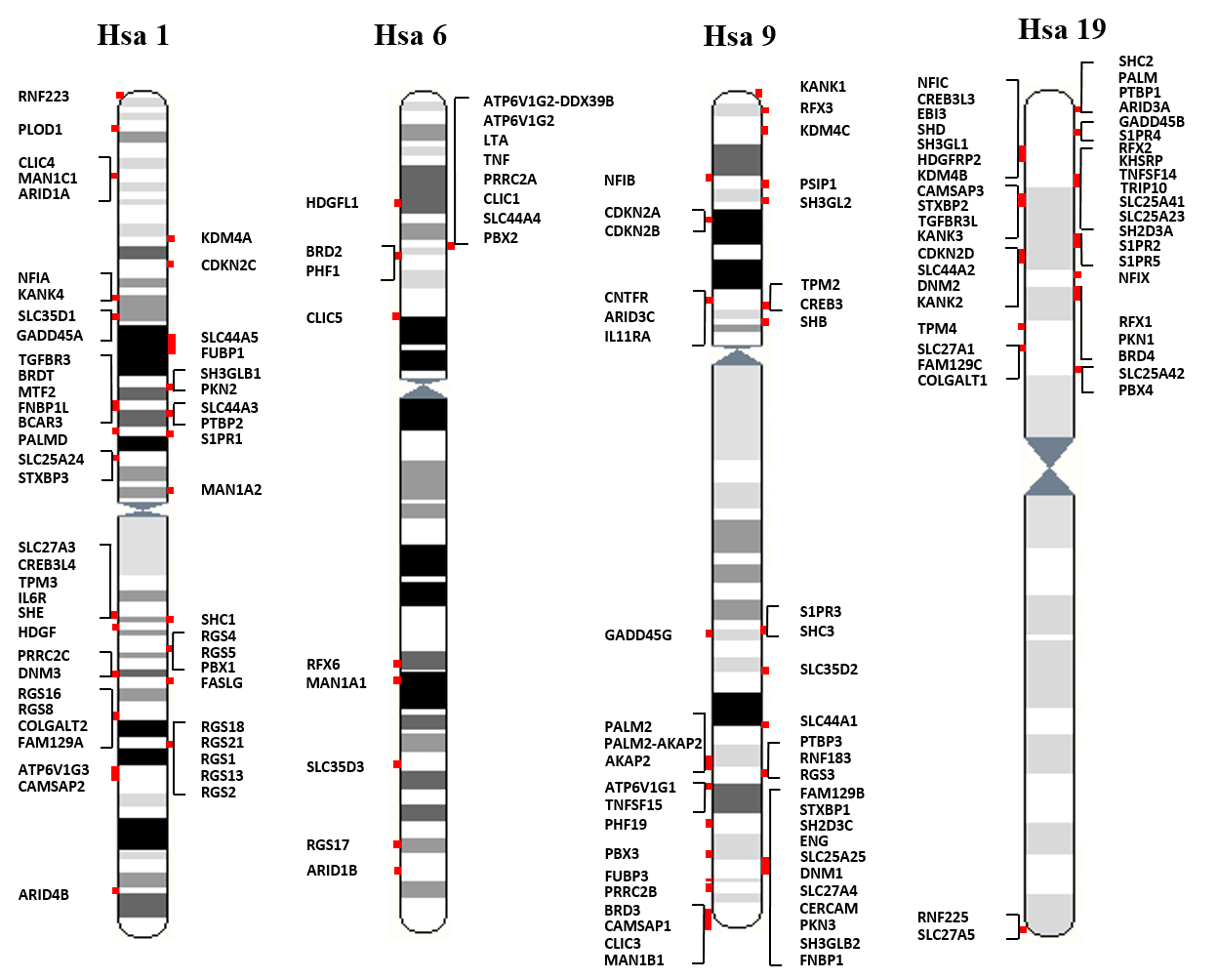
Figure 1: Gene families with members on at least three of the human MHC-bearing chromosomes 1, 6, 9, and 19.
Useful references:
Parveen N, Masood A, Iftikhar N, Minhas BF, Minhas R, Nawaz U, Abbasi AA (2013) Comparative genomics using teleost fish helps to systematically identify target gene bodies of functionally defined human enhancers. BioMed Central Genomics, 14, 122.
Ali NM, Tsuboi R, Matsumoto Y, Koishi D, Inoue K, Maeda K, Kurata H (2016) Web application for genetic modification flux with database to estimate metabolic fluxes of genetic mutants. Journal of bioscience and bioengineering, 122, 111.
Flicek, P., Amode, M.R., Barrell, D., Beal, K., Brent, S., Chen, Y., Clapham, P., Coates, G., Fairley, S., Fitzgerald, S., Gordon, L., Hendrix, M., Hourlier, T., Johnson, N., Kahari, A., Keefe, D., Keenan, S., Kinsella, R., Kokocinski, F., Kulesha, E., Larsson, P., Longden, I., McLaren, W., Overduin, B., Pritchard, B., Riat, H.S., Rios, D., Ritchie, G.R., Ruffier, M., Schuster, M., Sobral, D., Spudich, G., Tang, Y.A., Trevanion, S., Vandrovcova, J., Vilella, A.J., White, S., Wilder, S.P., Zadissa, A., Zamora, J., Aken, B.L., Birney, E., Cunningham, F., Dunham, I., Durbin, R., Fernandez-Suarez, X.M., Herrero, J., Hubbard, T.J., Parker, A., Proctor, G., Vogel, J., Searle, S.M., (2011) Ensembl 2011. Nucl. Acids Res, 39, D800–D806.
Fujita, P.A., Rhead, B., Zweig, A.S., Hinrichs, A.S., Karolchik, D., Cline, M.S., Goldman, M., Barber, G.P., Clawson, H., Coelho, A., Diekhans, M., Dreszer, T.R., Giardine, B.M., Harte, R.A., Hillman-Jackson, J., Hsu, F., Kirkup, V., Kuhn, R.M., Learned, K., Li, C.H., Meyer, L.R., Pohl, A., Raney, B.J., Rosenbloom, K.R., Smith, K.E., Haussler, D., Kent, W.J., (2011) The UCSC genome browser database: update 2011. Nucl. Acids Res, 39, D876–882.
Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., Durbin, R., Eyras, E., Gilbert, J., Hammond, M., Huminiecki, L., Kasprzyk, A., Lehvaslaiho, H., Lijnzaad, P., Melsopp, C., Mongin, E., Pettett, R., Pocock, M., Potter, S., Rust, A., Schmidt, E., Searle, S., Slater, G., Smith, J., Spooner, W., Stabenau, A., Stalker, J., Stupka, E., Ureta-Vidal, A., Vastrik, I., Clamp, M., (2002) The ensembl genome database project. Nucl. Acids Res, 30, 38–41.
