Synteny of HSA 1/2/8/20 Paralogon
The conservation of gene synteny on different chromosomal region entails the functional significance of these genes. For instance, the co-expression of neighboring genes is mediated by high order structural organization of chromosomes, which brings together the far off genomic regions in close proximity in order to be expressed together in a coordinated manner (Parveen et al. 2013). Similarly, the gene regulatory elements spread across long regions impose critical constraint on genomic architecture and are known to have maintained exceptionally long syntenic blocks both within and across species (Meaburn and Misteli 2007).
To investigate the evolutionary mechanisms that shaped the quadruplicated paralogy blocks residing on human chromosomes 1, 2, 8, 20, rigorous scanning of these chromosomes was performed. Thorough scanning identified 11 multigene families (40 genes), seven with fourfold while four with three fold representation on Hsa 1/2/8/20. Synteny map of the paralogy blocks depicting the actual order of genes was drafted and presented in Figure 1.
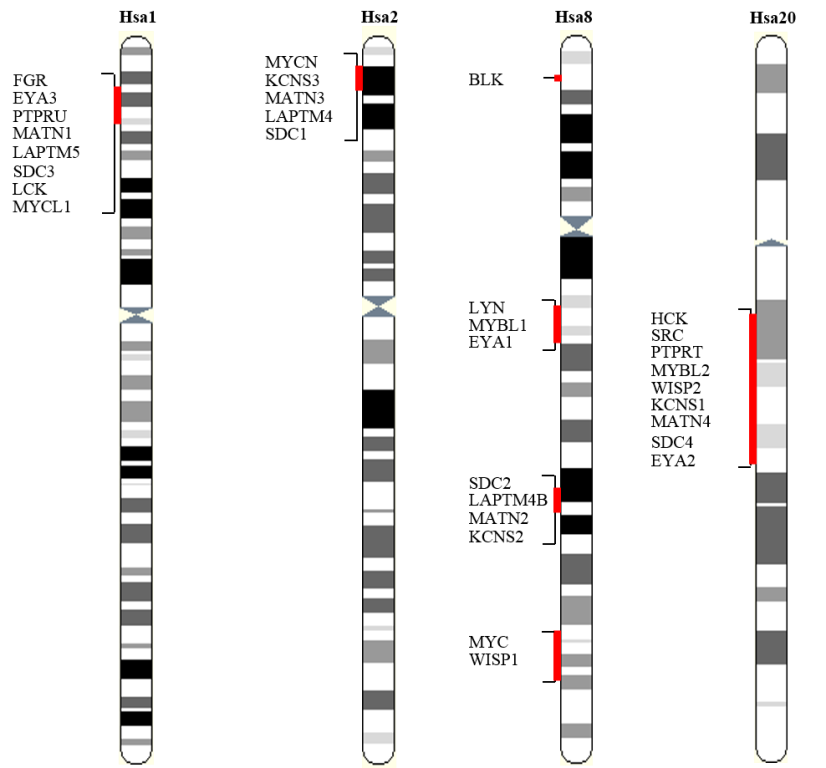
Figure 1: Gene families with members on at least three chromosomes of the human 1, 2, 8, and 20 paralogon.
Useful references: