Segmental Duplication
Duplications that encompass segments of DNA smaller than whole chromosomes are termed as segmental duplications (SDs). Segmental duplications are continuous portion of DNA that map two or more genomic location which can also defined as low copy repeats(LCRs) (Bailey et al. 2006). These duplications are in fact large blocks of genomic sequences ranging in size of >1kb in length and showing a sequence identity of more than 90% (Cheung et al. 2003). Segmental duplications arbitrate by cytogenetic alteration mechanisms including deletion, duplication and inversion (Emanuel et al. 2001) (Figure 1).
About 5.2% of human genome consists of SDs that has emerged during the past 35 million years of our species’ evolution (Bailey et al. 2002). Certain SDs expands and leads to birth of novel genes and regulatory elements through duplicative mechanism (Lynch et al. 2000).
Recent investigation suggest SDs have also occurred deep in vertebrate history or prior to fish tetrapod split. It has been proposed that such segmental duplication/Co duplication event play very critical role in early genome evolution (Abbasi 2010a; Abbasi 2010b; Ambreen S et al. 2014). These ancient SDs (prior to fish tetrapod split) have been termed as ancient segmental duplication (aSDs) to discriminate them from recent segmental duplication (rSDs) that have occurred during recent primate history (Ajmal et al. 2014; Abbasi et al. 2015).
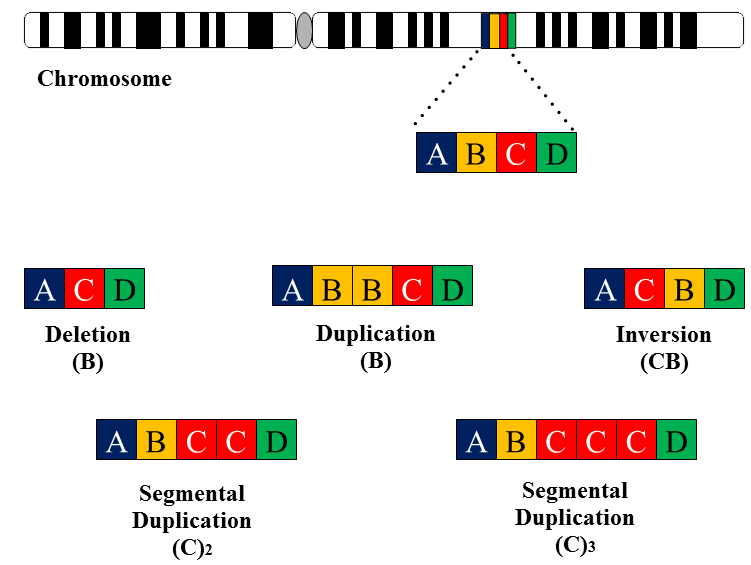
Figure 1: Segmental duplication occurs underneath mechanisms of gene deletion, insertion and duplication that could be inter chromosomal or intra chromosomal duplication. Duplication shadowing shows that unique region where existing duplication sponsor further duplications like (C) gene depicts duplication shadowing region.
Useful references:
Bailey JA, Eichler EE (2006) Primate segmental duplications: crucibles of evolution, diversity and disease. Nature Reviews Genetics, 7, 552.
Cheung, J., X. Estivill, R. Khaja, J. R. MacDonald, K. Lau, L. C. Tsui, and S. W. Scherer (2003) Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biology, 4:R25.
Beverly S. Emanuel and Tamim H. Shaikh (2001) Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nature Reviews Genetics, 2, 791.
Bailey, J. A., Gu, Z., Clark, R. A., Reinert, K., Samonte, R. V., Schwartz, S.& Eichler, E. E. (2002) Recent segmental duplications in the human genome. Science, 297, 1003-1007.
Michael Lynch and John S. Conery (2000) The evolutionary fate and consequences of duplicate genes. Science, 290, 1151.
Abbasi A.A.* (2010) Piecemeal or big bangs: correlating the vertebrate evolution with proposed models of gene expansion events . Nat Rev Genet, 11(2), 166.
Abbasi A.A.* (2010) Unraveling ancient segmental duplication events in human genome by phylogenetic analysis of multigene families residing on HOX cluster paralogons . Molecular Phylogenetics and Evolution, 57, 836-848.
Ambreen S , Khalil F , Abbasi A.A*. (2014) Integrating large-scale phylogenetic datasets to dissect the ancient evolutionary history of vertebrate genome . Molecular Phylogenetics and Evolution, 78, 1-13.
Ajmal, W., Khan, H., Abbasi, A.A*. (2014) Phylogenetic investigation of human FGFR-bearing paralogons favors piecemeal duplication theory of vertebrate genome evolution. Molecular Phylogenetics and Evolution, 81, 49-60.
Hafeez, M., Shabbir, M., Altaf, F., and Abbasi, A.A*. (2016) Phylogenomic analysis reveals ancient segmental duplications in the human genome. Molecular Phylogenetics and Evolution, 94, 95-100.
Samonte RV, Eichler EE (2002) Segmental duplications and the evolution of the primate genome. Nature Reviews Genetics, 3, 65.
