Age estimates of duplications: MHC Paralogon
The order of branching within the phylogenetic tree was used to estimate the time window of gene duplication events relative to the divergence of major taxa of organisms. This method of relative dating does not rely on the assumption of a constant rate of evolution. Therefore the process is sensitive to the varying rate of evolution in different branches of the tree (Hughes, 1998).
For 40 multigene families (in total 175 genes) with triplicated and quadruplicated distribution on Hsa 1/6/9/19, 28 duplication events were detected before vertebrate–invertebrate split and 102 duplications were detected after vertebrate–invertebrate and before tetrapod–bony fish divergence. Only 7 tetrapod specific duplication events were detected (Figure 1) (Naz et al., 2017)
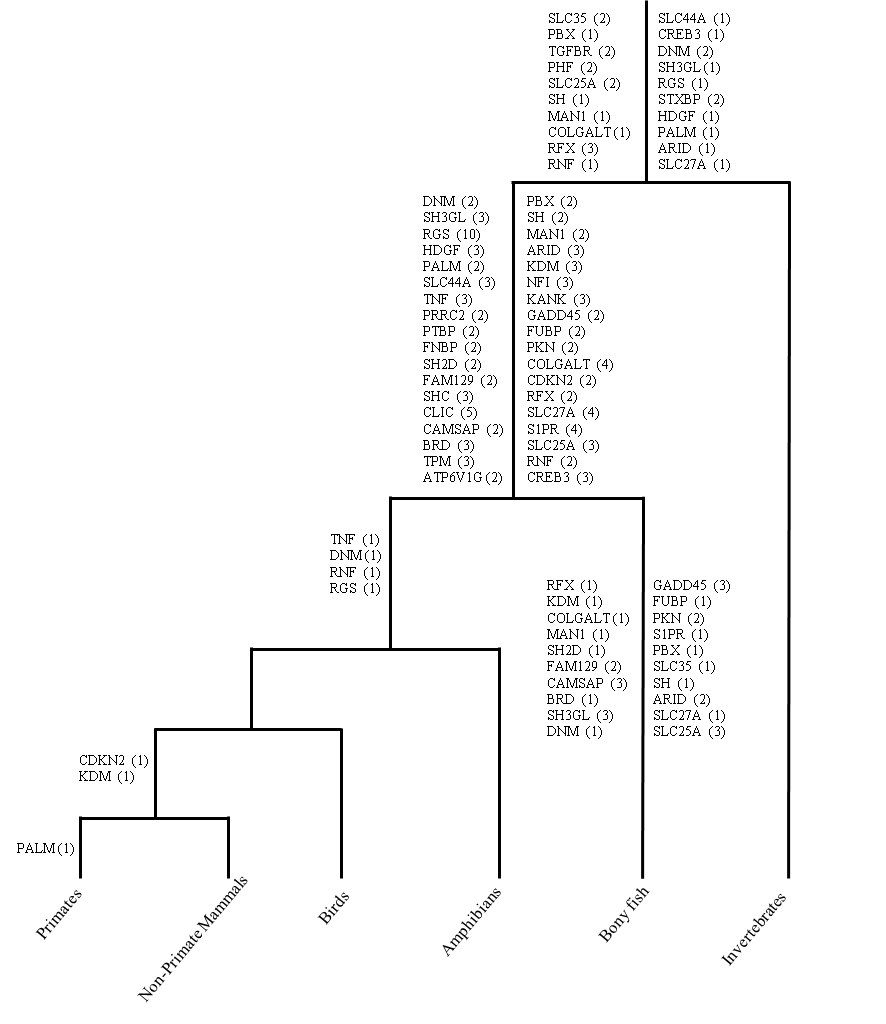
Figure 1: The relative timing of duplication events that expanded the multigene families residing on human MHC paralogon.
Useful references: