HOX (HSA 2/7/12/17) Paralogon Information
The axial patterning of all animal forms is patterned during early embryo-genesis (DE ferrier et al, 2001). Over the past couple of decades, evolutionary and developmental studies focussed extensively on elucidating semantic underpinning of pattern formation in bilateral life forms. Mutational studies of homeobox genes (HOX), a family of transcriptional factors that harbors a 60-amino acid homeodomain, provides most informative clue regarding development of main body area of bilaterians and during early embryonic development (J Garcia-Fernàndez, 2005).
Homologs of HOX genes are found in wide variety of animals, ranging from invertebrates to mammals. One group of HOX genes are clustered in close physical proximity and thus termed as “HOX cluster”. This unique clustered genomic organization is widespread across diverse animal forms and has functional significance (Duboule, 2007). All invertebrates including amphioxus possess at least single HOX cluster with maximum of 14 paralogs arranged in tandem, whereas mammalian genome possesses approximate 39 HOX genes arranged in 4 distinct clusters (Wolfe, 2001) (Figure 1). The collinear arrangement of HOX paralogues in a cluster often reflects spatial and temporal order of their expression and their function along oral-aboral axis (D Lemons et al, 2006). The HOX genes at 3' end are expressed earlier and pattern the anterior of the embryo, whereas genes at the 5' end of the cluster are expressed at a later developmental stage and pattern the posterior part of the body (Meyer A, 1998). Presence of intact HOX cluster across vertebrate-invertebrate lineage for over 900 million years indicates that this unique genomic organization is evolutionarily constrained. Over the past few years, it has become clear that the cis-regulatory elements spread across the HOX cluster containing chromosomal loci are responsible for the evolutionary inertia on HOX organization (J.C. Pearson et al, 2005).
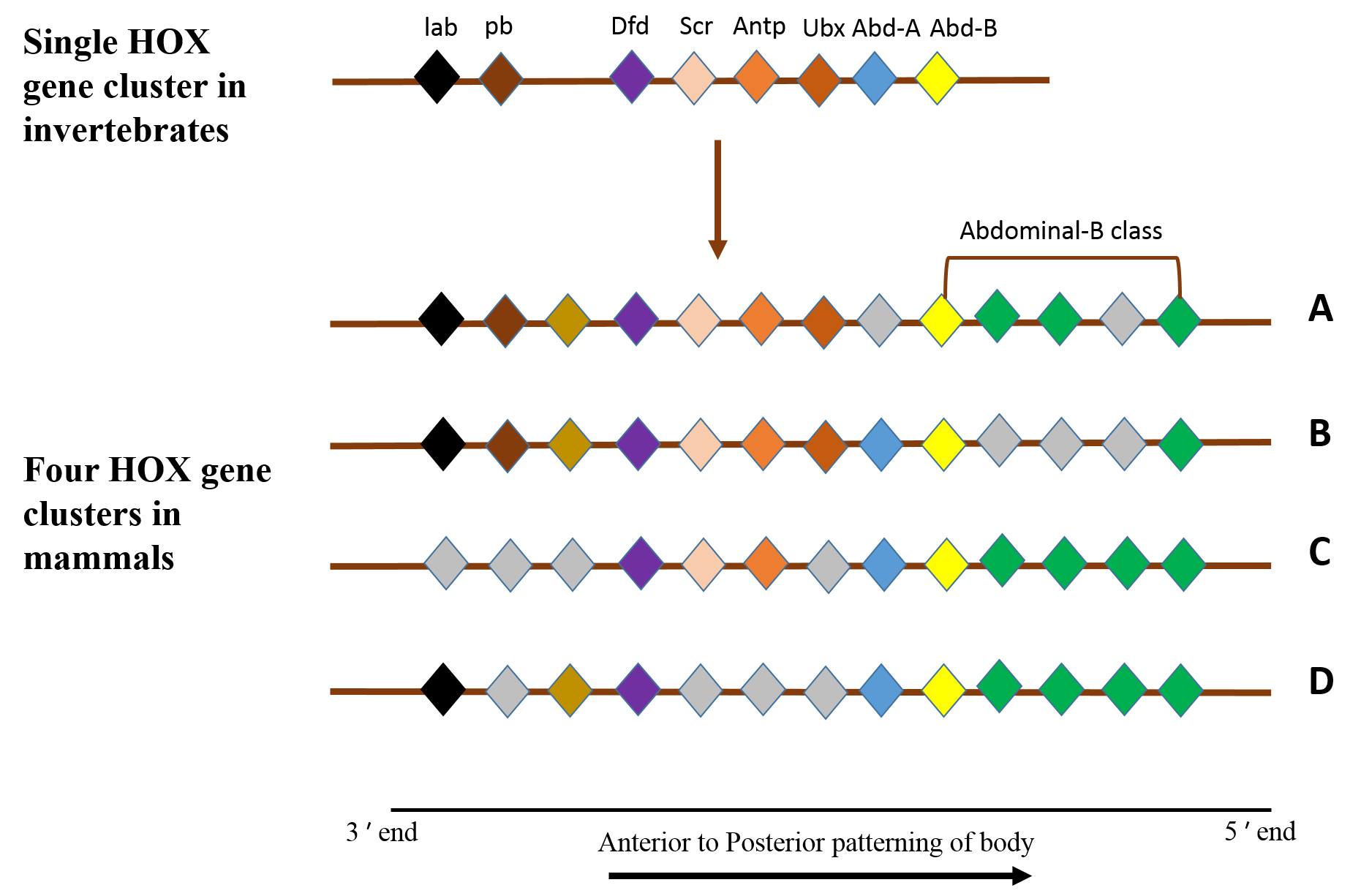 Figure 1: All invertebrates (including Amphioxus and Drosophila) possess single HOX gene cluster, whereas all vertebrtaes (including mammals) possess at least four HOX gene clusters. Genes are colored to differentiate between Hox family members, and genes that are orthologous between clusters and species are labeled in the same color. Genes are shown in the order in which they are found on the chromosomes but, for clarity, some non-Hox genes that are located within the clusters are colored Grey.
Figure 1: All invertebrates (including Amphioxus and Drosophila) possess single HOX gene cluster, whereas all vertebrtaes (including mammals) possess at least four HOX gene clusters. Genes are colored to differentiate between Hox family members, and genes that are orthologous between clusters and species are labeled in the same color. Genes are shown in the order in which they are found on the chromosomes but, for clarity, some non-Hox genes that are located within the clusters are colored Grey.
Intra-genomic comparison of mammalian genomes identified several potentially triplicated and quadruplicated syntenic segments, termed as paralogons. One such paralogon in human genome famously known as HOX cluster paralogon contains various distinct multigene families in triplicated and quadruplicated arrangement around four coherent human HOX clusters. This database provides robust and thorough phylogenetic investigation of at least 62 multigene families residing in triplicated or quadruplicated configuration on human HOX cluster bearing paralogon. This large scale data in turn provides an insight into ancient mechanism (prior to fish-tetrapod split) shaping the current architecture of human chromosome 2, 7, 12, and 17.
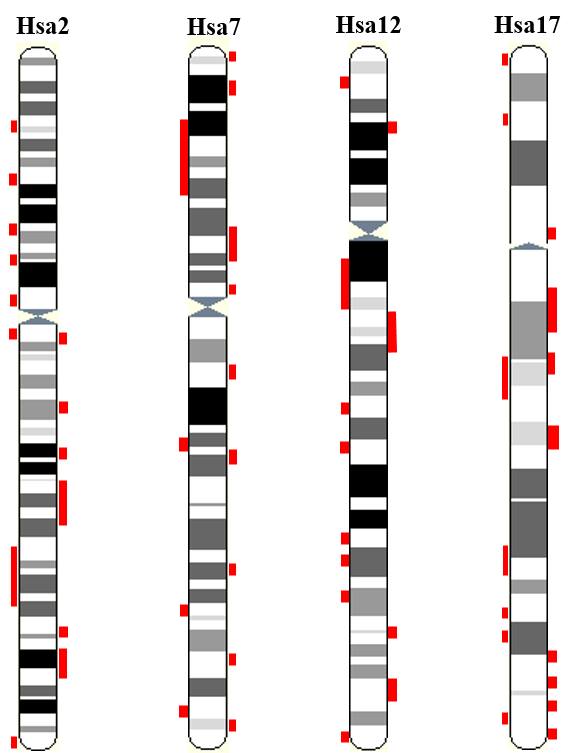 Figure 2: Paralogy region residing on human HOX-bearing chromsomes 2, 7, 12 and 17 are highlighted in red.
Figure 2: Paralogy region residing on human HOX-bearing chromsomes 2, 7, 12 and 17 are highlighted in red.
Useful references: