FGFR (HSA 4/5/8/10) Paralogon Information
Fibroblast growth factors are polypeptide growth factors with varied biological actions. Fibroblast growth factors (FGFs) and their receptors regulate developmental processes such as mesoderm formation and gastrulation. Fibroblast growth factor receptors (FGFRs) mediate a wide spectrum of cellular responses that are crucial for development and wound healing (A, Guilio et al., 2013). Furthermore, they are also involved in the growth of various tissues like ear, limb, hair and skeletal systems (Mason, 1994; OM Wilkie et al., 1995; Yamaguchi and Rossant, 1995). Interaction of FGF ligands with their signaling receptors is regulated by protein or proteoglycan cofactors and by extracellular binding proteins (DM Ornitz et al., 2015). Consistent with the presence of FGFs in almost all tissues and organs, aberrant activity of the pathway is associated with developmental defects that disrupt organogenesis, impair the response to injury, and result in metabolic disorders and cancer.
Human FGFs are comprised of a conserved core of ~120 amino acids, with a sequence identity of 30-60% leading to an overall homology of 56-71% among FGF genes (A Vienne. et al., 2003). Evolutionary studies of FGF gene families indicated the expansion of family in two phases resulting in 22 members which are grouped in eight subfamilies (Oulion et al., 2012). 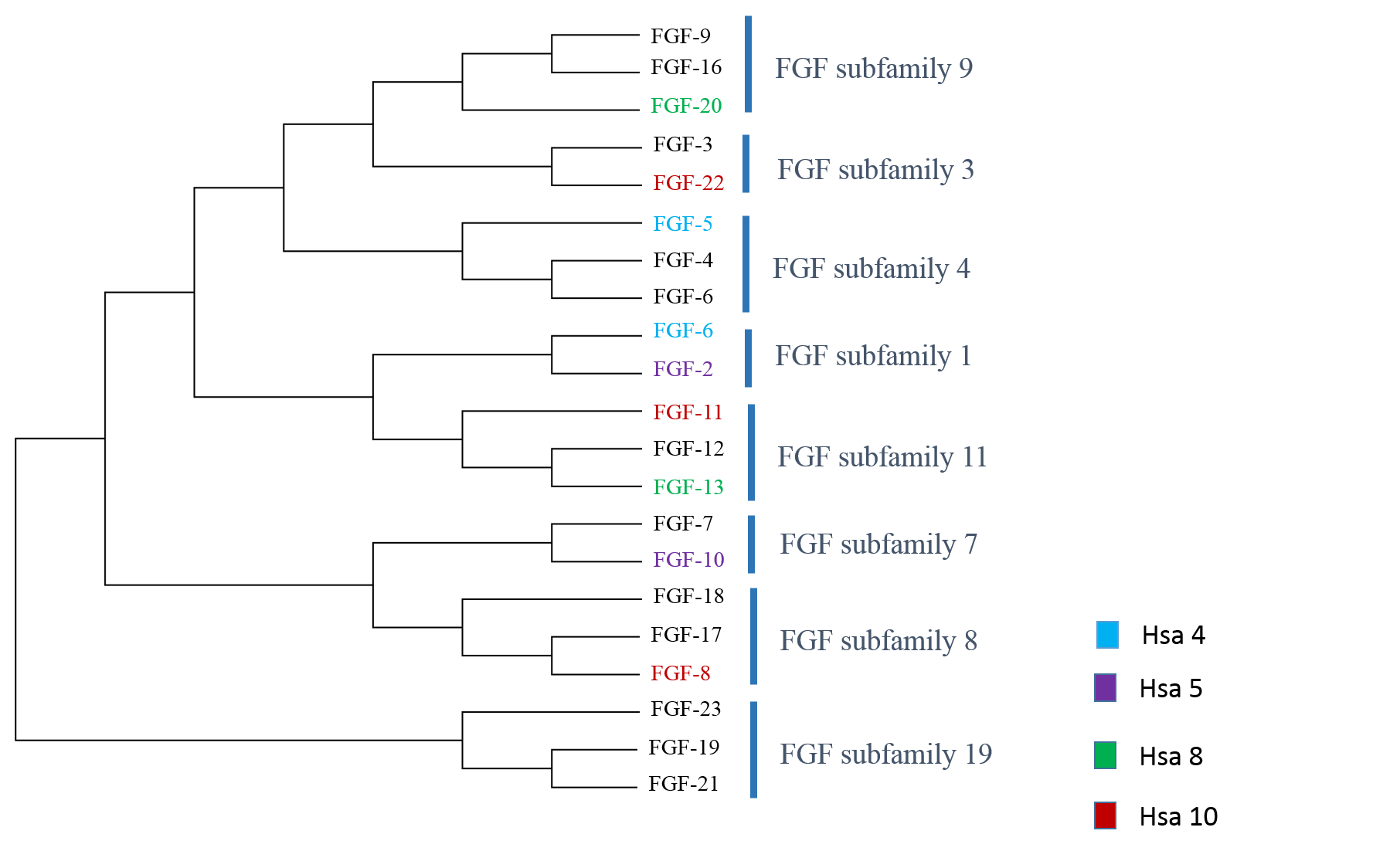
Figure 1: Classification of FGF genes on chromosomes 4, 5, 8 and 10. Members of gene families residing on each chromosome of the paralogon are colored to differentiate.
Phylogenetic analysis of FGFR genes shows that the four members of this family diversified early in vertebrate history. However, alternative splicing by FGFRs during the course of evolution has increased their functional diversity (D. M. Ornitz et al., 2004). The four members of FGFR are located on each chromosome on Hsa 4/5/8/10 and give it a name FGFR paralogon.
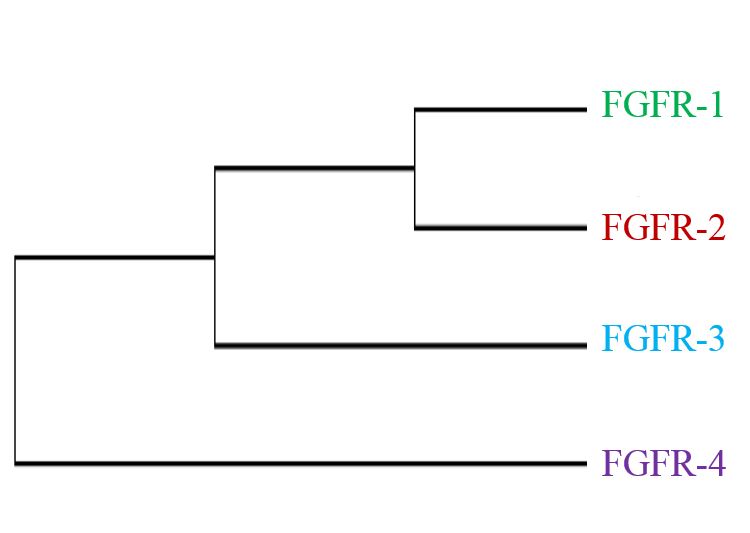
Figure 1: Classification of FGFR genes on chromosomes 4, 5, 8 and 10. Members of gene families residing on each chromosome of the paralogon are colored to differentiate.
It was also speculated that FGF and HOX gene families might have coevolved and thus contributed in conjunction towards augmented complexity in vertebrates (Coulier et al., 1997). Human FGFR bearing paralogon consists of 80 multigene families; 17 with four fold representation while 63 with three fold representation on Hsa 4/5/8/10.
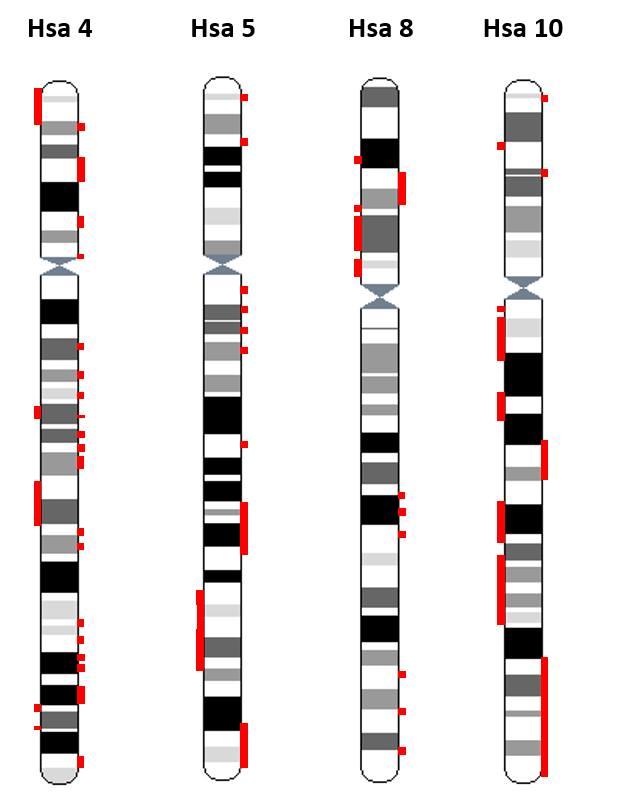
Figure 3: Paralogy regions residing on human FGFR-bearing chromosomes (Hsa 4/5/8/10) are highlighted red.
Useful references: