FGFR paralogon Co-duplicated groups
It was proposed that elucidation of intra-genomic syntenic regions through map self-comparison approach does not provide compelling support for the proposed mechanism of origin of paralogons (Abbasi, 2008). To present a plausible explanation of the evolutionary events that shaped the syntenic relationships seen on present day human FGFR bearing chromosomes, topology comparison approach was employed for triplicated/quadruplicated gene families residing on this paralogon (Abbasi, 2010b; Abbasi and Grzeschik, 2007; Zhang and Nei, 1996). Gene families belonging to a particular co-duplicated group suggest that they share similar evolutionary history and might have originated through gene cluster duplication event, whereas genes belonging to different co-duplicated groups may not share their evolutionary history and might not have duplicated simultaneously. Duplication history summary of families residing on FGFR paralogon with respect to speciation events and identified co-duplication events are given in the following table.
| Chromosomes/Paralogon | ||||||||
| Name of Co-duplicated group | Co-duplicated Group A | Co-duplicated Group B | Co-duplicated Group C | Co-duplicated Group D | Co-duplicated Group E | Remaining Families | Largely duplicated in invertebrates (prior to vertebrate invertebrate split) | Total |
| No. of gene families | 6 | 6 | 11 | 18 | 10 | 24 | 8 | 82 |
For 82 multigene families residing on FGFR bearing paralogon (HSA 4/5/8/10), 5 distinct co-duplicated groups were identified with gene families diversified within the time window of vertebrate-invertebrate and tetrapod-bony fish split. First co-duplicated group presented a topology of type ((Hsa4/Hsa2 Hsa5)(Hsa8/Hsa2 Hsa10)) and suggests the simultaneous duplication of 6 gene families (Figure 1-A). Second co-duplicated group comprised members from 6 gene families and shows a topology of type ((Hsa5 Hsa10)Hsa4/Hsa8)(Figure 1-B). Third co-duplicated group included 11 gene families and shows a topology of type ((Hsa4 Hsa10/Hsa2)Hsa5/Hsa8)(Figure 1-C). Fourth co-duplicated group was the largest with 18 gene families and follows a topology of type (((Hsa4/Hsa20 Hsa5/Hsa2) Hsa10/Hsa11) Hsa8/Hsa2)(Figure 1-D). Fifth co-duplicated group shows a topology of type (((Hsa8 Hsa10) Hsa5/Hsa20) Hsa4/Hsa5) and involved gene members from 10 families (Figure 1-E)(Hafeez et al, 2016).
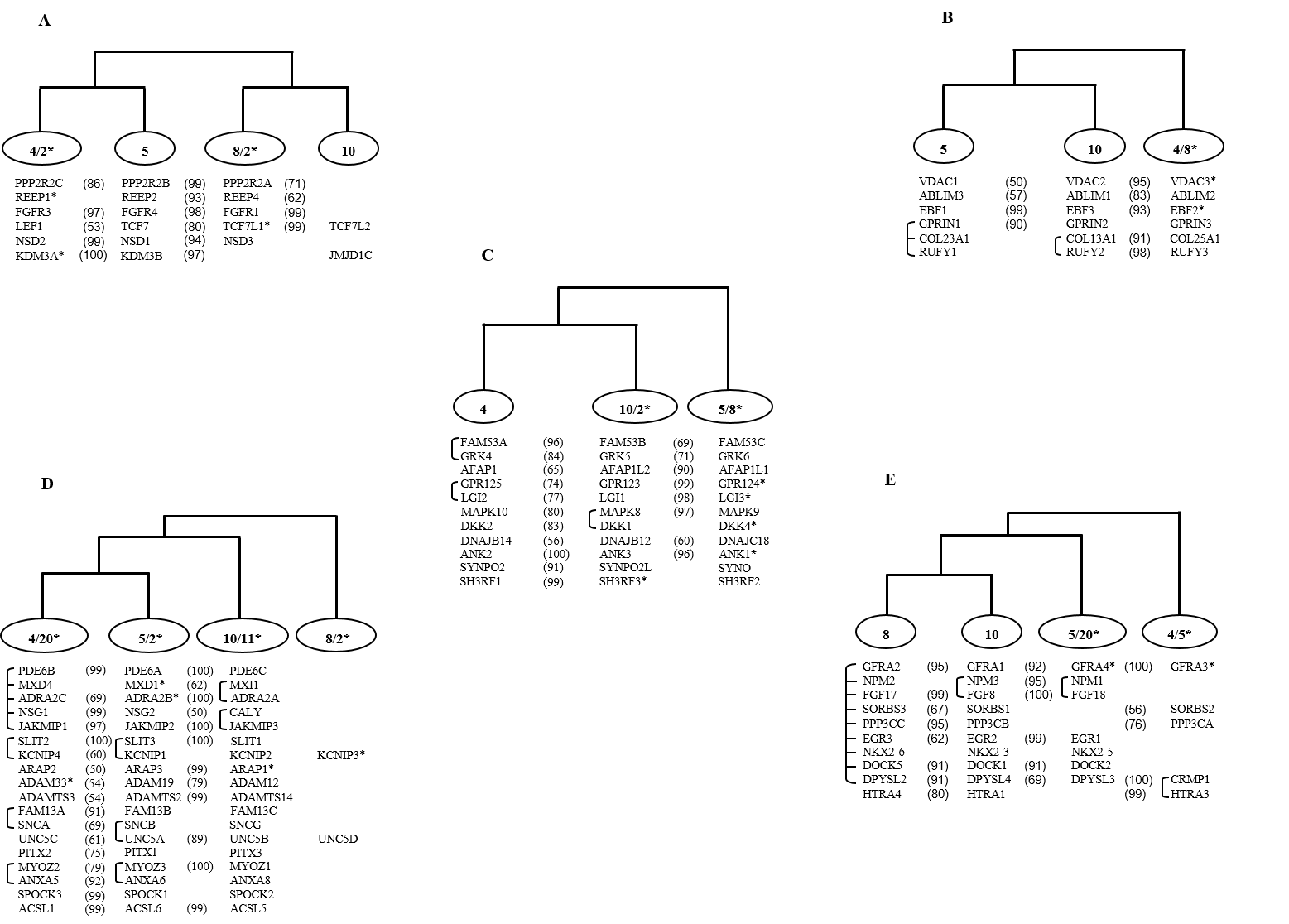
Figure 1: Schematic topologies indicating consistencies in phylogenies of families including members on at least three of the human 4, 5, 8, 10 chromosomes. In each case the percentage bootstrap support is given in parentheses and only values ≥50% are presented here. The connecting bars on the left depict the close physical linkage. * Represents situation where a gene family member is not residing on FGFR bearing chromosomes.
Useful references: