HSA1/2/8/20 Paralogon Information
The multiple sets of collinear triplicates and quadruplicates known as paralogons present on several human chromosomes are considered as a characteristic spectacle towards the evolution of eukaryotic genome.
Thorough investigation of the human genome identifies several clusters of genes with two to four fold representations on the chromosomes. Most investigated and famously known clusters include HOX cluster (Hsa 2/7/12/17) and FGFR (Hsa 4/5/8/10) clusters. In addition to these, there exist multiple other triplicated/quadruplicated paralogons in the human genome. For instance the linkage groups located on human chromosomes 1, 2, 8 and 20 were investigated both in contest of small and large scale vertebrate gene/genome duplications (Gibson and Spring, 2000; Abbasi, 2012).
This database provides history of 11 triplicated and quadruplicated gene families residing on Hsa 1/2/8/20 paralogon (Figure 1)(Abbasi, 2012). Of these 11 multigene families, seven showed a fourfold paralogy on these chromosomes while four gene families exhibited three fold representation on the paralogon.
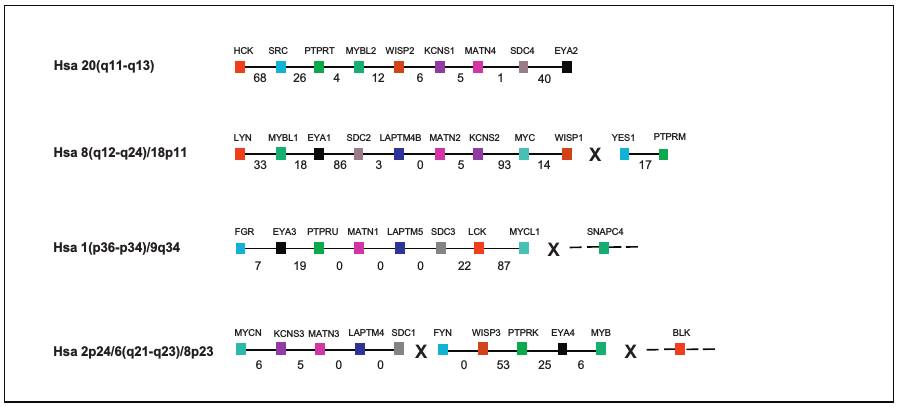
Figure 1: Schematic representation of paralogous genes residing in the proposed fourfold paralogy regions on Hsa 1/2/8/20.
This database provides robust and thorough phylogenetic investigation of multigene families residing in triplicated or quadruplicated configuration on human 1/2/8/20 paralogon. This large scale data in turn provides an insight into ancient mechanism (prior to fish-tetrapod split) shaping the current architecture of human chromosomes 1, 2, 8 and 20.
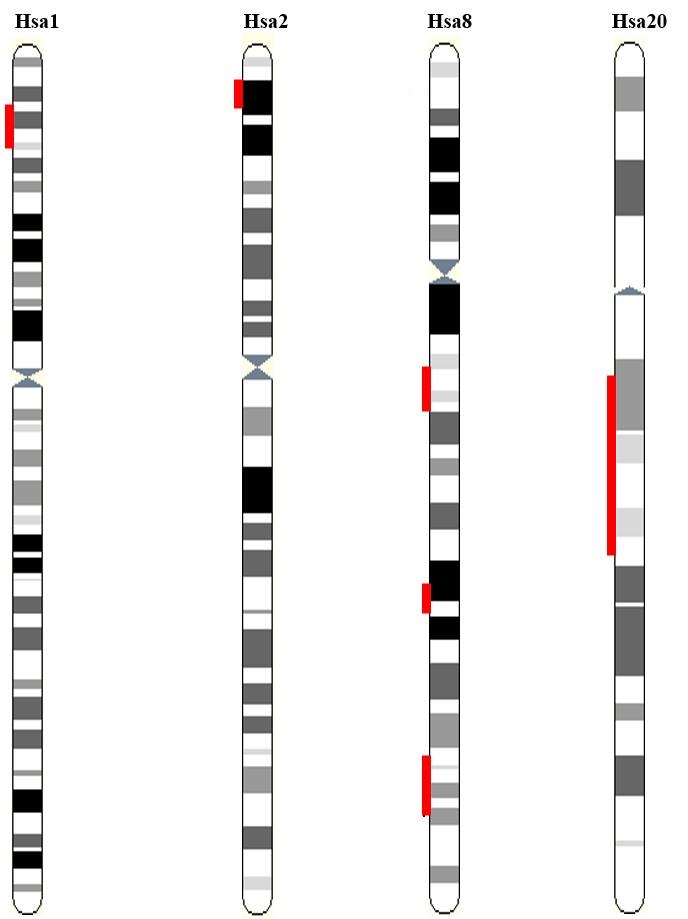
Figure 2: Triplicated and quadruplicated paralogy regions residing on Human Hsa 1/2/8/20 are highlighted red.
Useful references: