Paralogy Regions
Repertoires of paralogy groups are scattered over the entire human genome which are considered as the remnants of large scale gene/segment/genome duplication events (Ohno, 1970). In particular, presence of triplicated/quadruplicated human paralogy blocks such as Hsa 2/7/12/17 (HOX cluster), Hsa 4/5/8/10 (FGFR cluster), Hsa 1/6/9/19 (MHC cluster) and Hsa 1/2/8/20 are speculated to have shaped up by ancient two rounds of whole genome duplication events.
Alternative hypothesis suggests that the constituent multigene families of these anciently conserved vertebrate paralogy regions arose as a result of small scale duplications involving gene clusters and chromosomal segments which occurred at different time points during chordate evolution (Furlong and Holland, 2002). Whereas the spread of similar gene sets on distinct vertebrate chromosomes is the consequence of subsequent rearrangement events. The conservation of these blocks of genes both within and between species may reflect a similar expression or function of these genes in the physical proximity on a chromosomal segment (Abbasi, 2008).
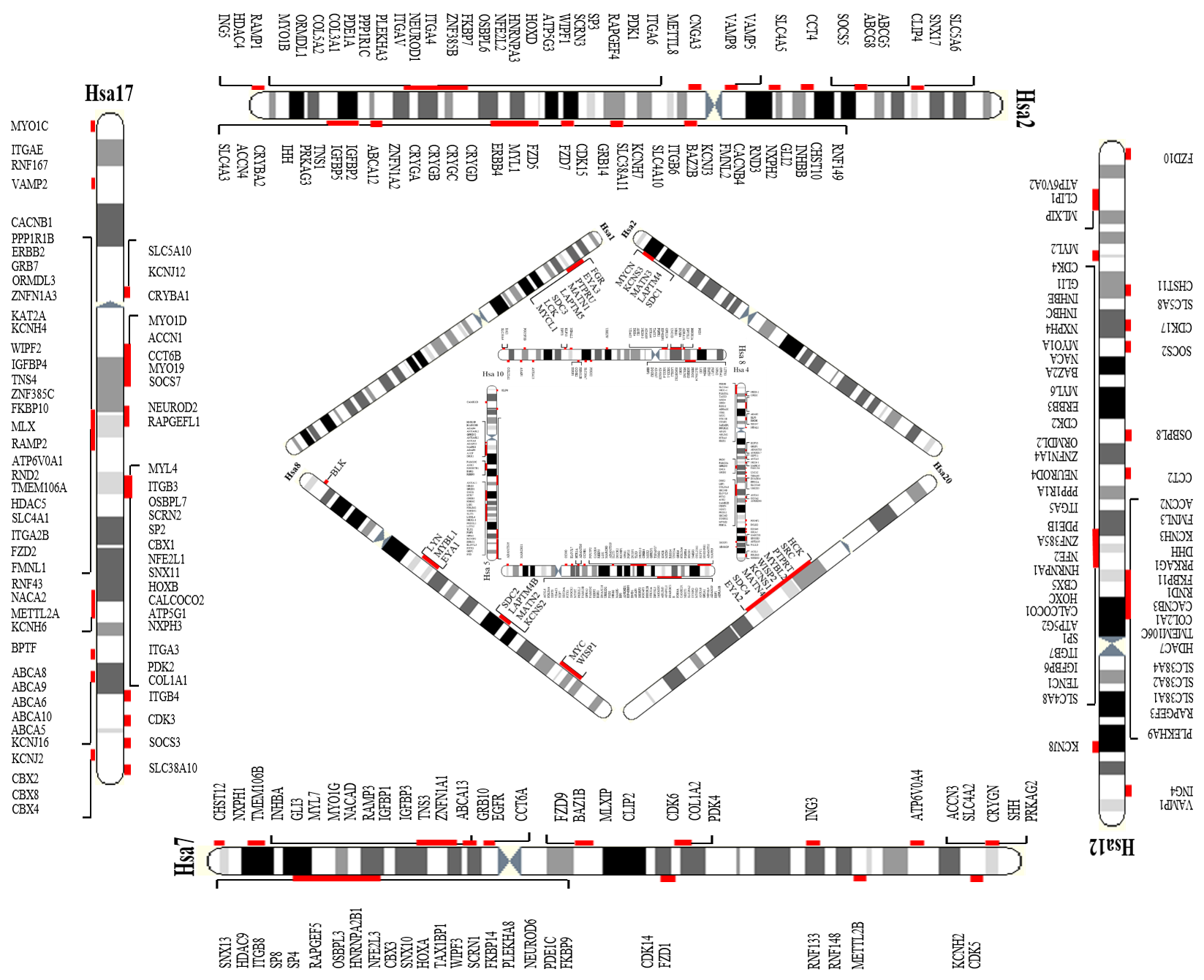
Figure 1: Four of the paralogy blocks in human genome i.e Hsa 2/7/12/17, Hsa 4/5/8/10, Hsa 1/2/8/2/20 and Hsa 1/6/9/19 are presented in this picture. Triplicated/quadruplicated gene families residing on these paralogons are highlighted red in this figure.
Useful references: