Co-duplication Events
It was proposed that elucidation of intra-genomic syntenic regions through map self-comparison approach does not provide compelling support for the proposed mechanism of origin of paralogons (Abbasi, 2008). To present a plausible explanation of the evolutionary events that shaped the syntenic relationships seen on the paralogons: Hsa 1/2/8/20, Hsa 2/7/12/17, Hsa 4/5/8/10 and Hsa 1/6/9/19, topology comparison approach was employed for gene families with triplicated or quadruplicated representatives on these paralogons. Branching order of phylogenetic trees is the key process to estimate the origin of paralogons. From given phylogenies only those gene families are evaluated which have been diversified within the time window of vertebrates-invertebrates and tetrapods-teleosts split, to find out which genes might have duplicated concurrently. Correspondence among the topologies of distinct multigene families comprising human paralogons would suggest the existence of simultaneous block or segmental duplications (Abbasi, 2010a; Abbasi, 2010b; Abbasi and Grzeschik, 2007; Abbasi and Hanif, 2012; Hughes et al., 2001). Gene families share the evolutionary history and have duplicated in accordance with each other if they belong to the same co-duplicated group, while the genes belonging to different groups have dissimilar evolutionary histories and follow different timings for duplication events. Duplication history summary of families with respect to speciation events and identified coduplication events are given in the following table.
| Chromosomes/Paralogon | |||||||
| Name of Co-duplicated group | Co-duplicated Group A | Co-duplicated Group B | Co-duplicated Group C | Co-duplicated Group D | Remaining Families | Largely duplicated in invertebrates (prior to vertebrate invertebrate split) | Total |
| No. of gene families | 12 | 6 | 4 | 9 | 24 | 8 | 62 |
| Chromosomes/Paralogon | ||||||||
| Name of Co-duplicated group | Co-duplicated Group A | Co-duplicated Group B | Co-duplicated Group C | Co-duplicated Group D | Co-duplicated Group E | Remaining Families | Largely duplicated in invertebrates (prior to vertebrate invertebrate split) | Total |
| No. of gene families | 6 | 6 | 11 | 18 | 10 | 24 | 8 | 82 |
| Chromosomes/Paralogon | |||||||||
| Name of Co-duplicated group | Co-duplicated Group A | Co-duplicated Group B | Co-duplicated Group C | Co-duplicated Group D | Co-duplicated Group E | Co-duplicated Group F | Remaining Families | Largely duplicated in invertebrates (prior to vertebrate invertebrate split) | Total |
| No. of gene families | 3 | 2 | 13 | 9 | 5 | 2 | 3 | 4 | 40 |
| Chromosomes/Paralogon | |||||||
| Name of Co-duplicated group | Co-duplicated Group A | Co-duplicated Group B | Co-duplicated Group C | Co-duplicated Group D | Remaining Families | Largely duplicated in invertebrates (prior to vertebrate invertebrate split) | Total |
| No. of gene families | 2 | 3 | 6 | 6 | 9 | 10 | 36 |
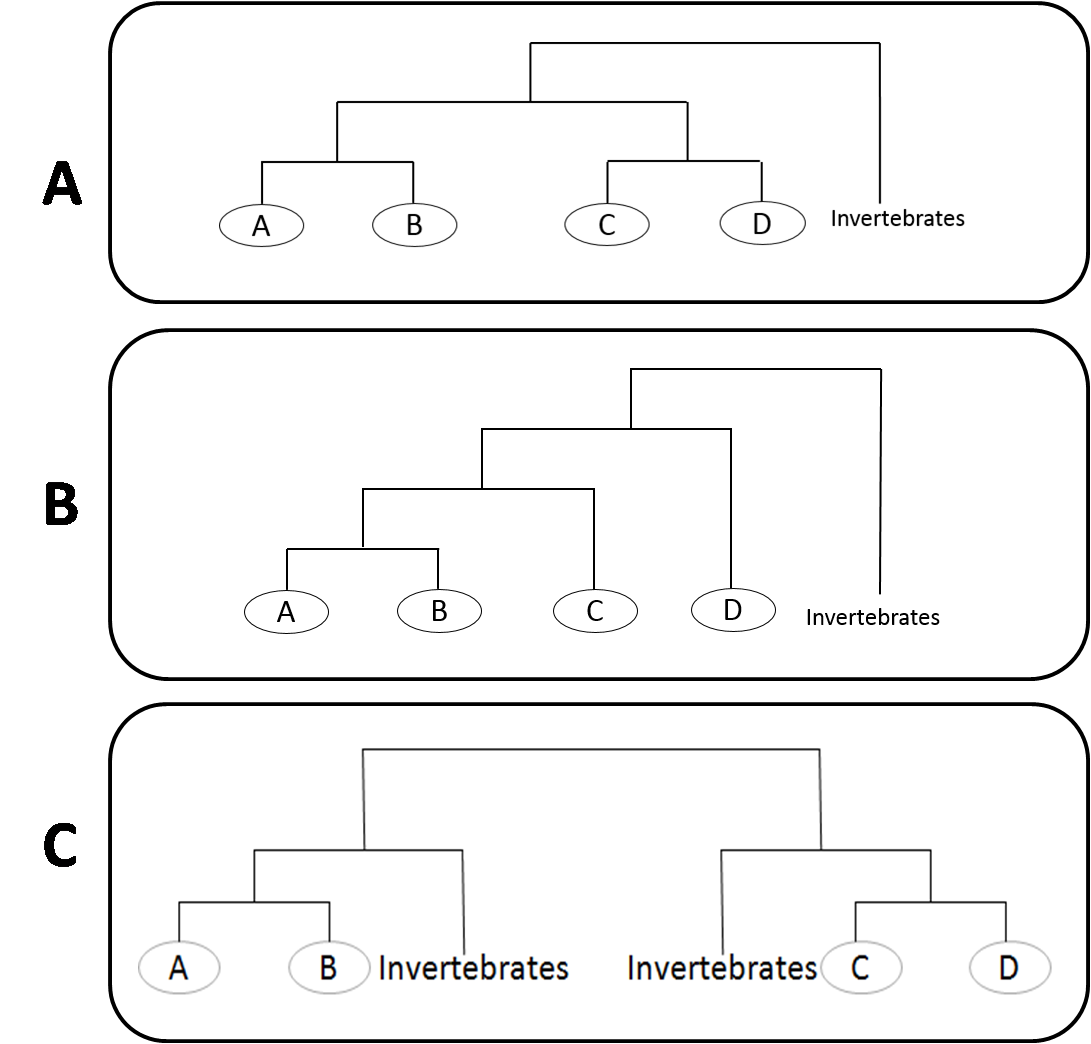
Under the 2R assumption, the vertebrate quadruplicated gene families should exhibit a symmetric tree topology showing two clusters of two paralogs with invertebrates falling outside the cluster, referred as ((AB)(CD)invertebrates) (Figure 1:A) (Hughes, 1998; Martin, 2001). Asymmetrical topology of the form ((((AB)C)D)invertebrates) shows independent gene duplications during vertebrate history (Figure 1:B)(Abbasi, 2010a; Hughes et al., 2001). Whereas (((AB)invertebrates)((CD)invertebrates)) depicts ancient origin prior to vertebrate-invertebrate split (Figure 1:B).
Our dataset presents the phylogenetic history of total 221 human gene families. Among these families in total 133 families appeared to have been originated through co-duplication events after vertebrate-invertebrate split but prior to fish-tetrapod split.
Figure 1: (A) Symmetric tree topology referred as ((AB)(CD)invertebrates), (B) Asymmetric tree topology referred as (((AB)C)invertebrates) and (C) Asymmetric tree topology referred as (((AB)invertebrates)((CD)invertebrates)).
Useful references: