Age Estimates of Duplications
A substantial way to determine the mechanistic of embarkation of several paralogy blocks on human chromosomes is by estimating the phylogenetic history of multigene families constituting these paralogons (Hughes, 1998; Hughes et al., 2001; Abbasi and Grzeschik, 2007). Relative timing approach helps to elucidate the time window and nature of ancient duplication mechanisms by estimating whether duplications have occurred prior or after a speciation event. Identifying the timing of duplication events relative to radiation of taxa is critical to accurate and adequate cross-species comparisons. Major evolutionary scenarios that prompted novelties and explosion in gene number throughout animal genomes can be taken in view by relative dating of these events. The method of relative dating does not rely on the assumption of a constant rate of evolution. Therefore the process is sensitive to the varying rate of evolution in different branches of the tree (Hughes 1998).
Among 193 triplicated/quadruplicated human families residing on HOX bearing paralogon (Hsa 2/7/12/17), FGFR bearing paralogon (Hsa 4/5/8/10), MHC bearing paralogon (Hsa 1/6/9/19) and Hsa 1/2/8/20 Paralogon, total 1179 duplications were spotted. Among these duplications, 351 duplication events were detected before vertebrate–invertebrate split. 588 duplications were detected after vertebrate–invertebrate and before tetrapod–bony fish divergence. Fish lineage showed 198 duplication events whereas only eight duplication events were detected at the root of tetrapod lineage. 11 primate specific duplications were detected while 120 duplications were spotted in the common ancestor of mammals. This analysis reveals that the current architecture of human genome is shaped by duplication events that are scattered across the entire history of animal lineage (Ambreen et al., 2014).
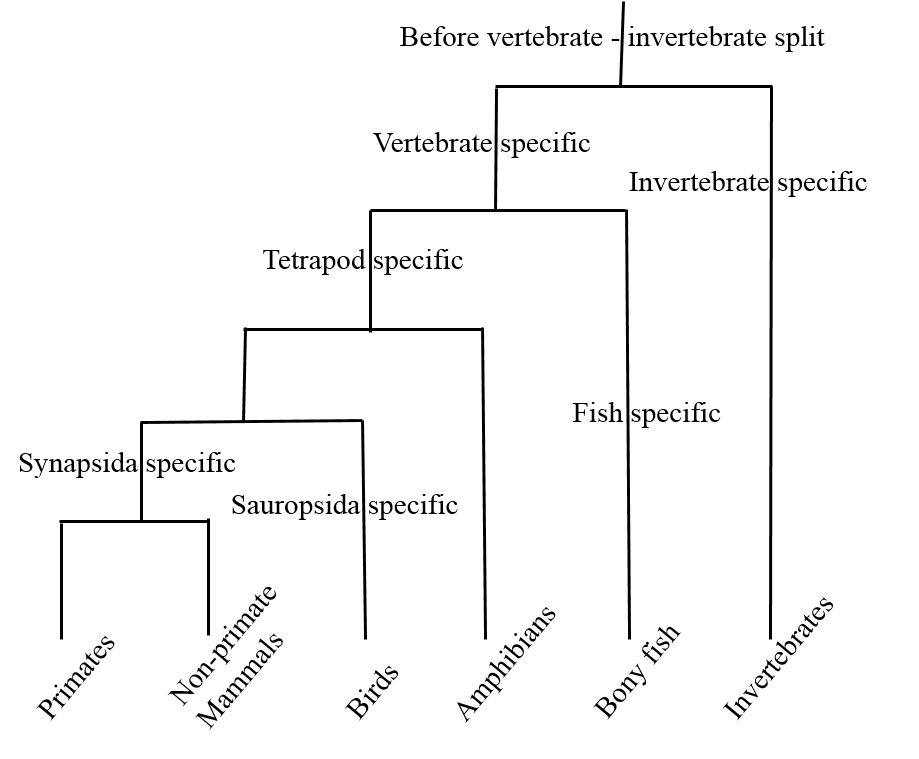
Figure 1: Generalized scheme depicting relative dating (relative to cladogenesis) of gene duplication events.
Useful references: